Ross Mounce is a postdoctoral researcher at the University of Bath studying the use of fossils in phylogeny and phyloinformatics, completing his PhD at the University of Bath last year. Ross was one of the first Panton Fellows and is an active member of the Open Knowledge Foundation, particularly the Open Science Working Group. He is an advocate for open science, and he is actively working on content mining academic publications to reuse scientific research in meta-analyses to gain higher level insights in evolutionary patterns.
Read more in my interview with him for Careers in Open Source Week.

Can you give us a brief overview of your research?
My current area of research is phyloinformatics, and I'm a postdoc in the Wills group at the University of Bath. I take published evolutionary trees and other evolutionary data from the academic literature and perform meta-analyses and syntheses of this information across hundreds and thousands of papers to gain higher-level insights into evolutionary patterns across different species groups. Just getting these data back into re-usable, re-computable forms from the published literature is by far and away the hardest challenge of our project. As part of the BBSRC-funded PLUTo project (Phyloinformatic Literature Unlocking Tools), I'm working with Peter Murray-Rust and the ContentMine team to develop software tools and approaches to help automate the process of finding and extracting phylogenetic data from the literature.
It's partly a needle in the haystack problem; there are 100,000+ papers published containing-phylogeny in the past decade, scattered across 1,000+ journals, and there are 2,000,000+ articles published per year!
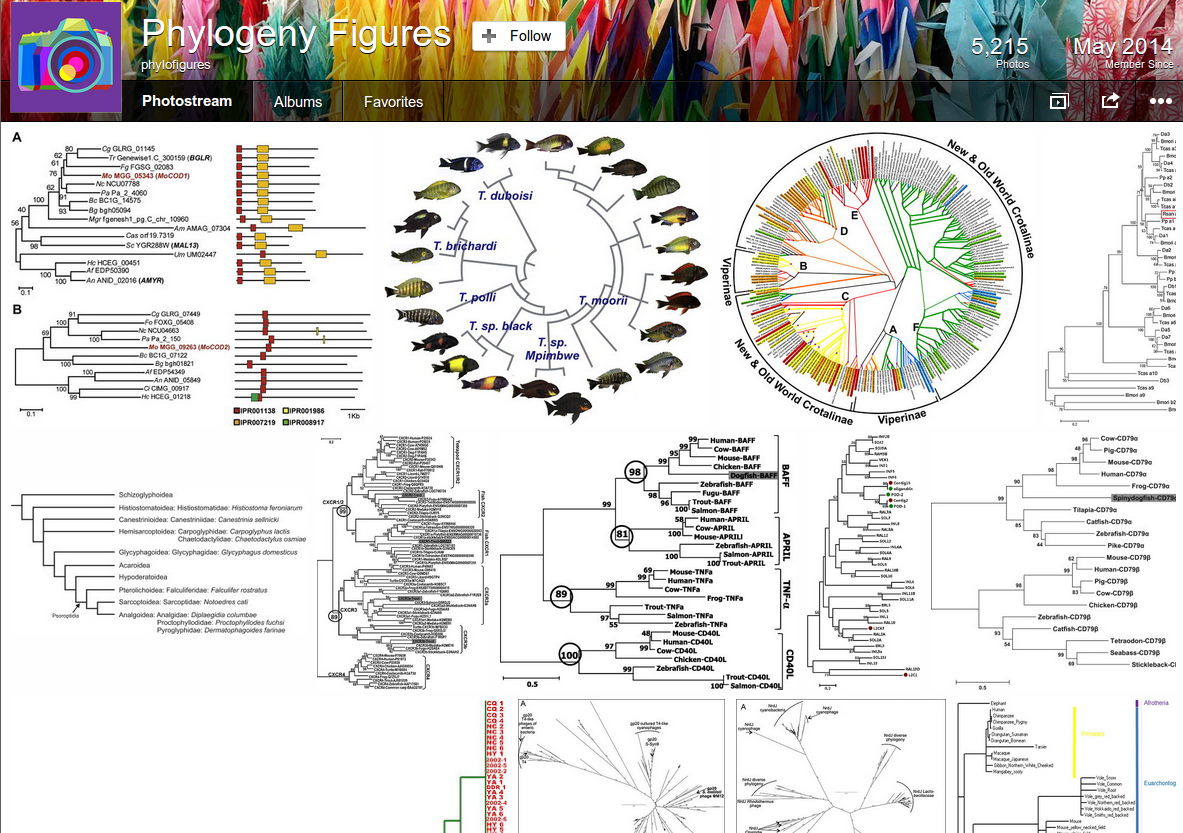
At the University of Bath, we don't even have legal access to all the journals in which we know phylogenetic data lies. Once found, data must typically be re-interpreted from the figure images provided in the publication. Only ~4% of published studies containing a phylogenetic analysis in 2010 provided machine-readable, re-usable data of their results. This 'data-poor' situation is not uncommon in many areas of science and is facilitated by the legacy journal publication system—most journals simply don't have strong data sharing requirements yet.
Why is open science, open source, and open data important to you?
Open science is vitally important to accelerating the pace of discovery and the continued funding of academic research. At least 80% of academic research is publicly, or charitably funded. It's therefore obvious that research should be done in a manner that maximizes the return-on-investment; encouraging sharing, re-use, and collaboration for overall gain. In 'closed' science, fewer people can read the publication (it's paywalled) and no one outside of the original author group can re-use the data or the code used to generate the results. The closed science model leads to deeply inefficient, slower, harder, progress. Researchers may overlook their peers papers simply because they don't have access to them. Likewise, researchers waste immense time and resources re-generating the same data or software functionality because other researchers didn't/won't share the original data/code.
Under the open science model, the publications are open for everyone to read and discover, and likewise the data and code are open for immediate re-use by all others too. It's clear to me that science would progress more quickly if it operated more frequently under the open model.
On a personal level, open science matters a great deal to me. I spent most of my PhD research time scraping data out of academic PDFs, or emailing authors (with relatively few helpful replies) for a copy of their published data. It was immensely frustrating. Instead of doing 'science' I was doing tedious, repetitive but highly-manual simple tasks. If authors had published their data alongside their papers according to long-established data formats I could have spent my time more usefully on re-analysis and extending the limits of our knowledge. I talked with my peers and found they had these problems too; the immense inefficiency was somehow 'normal' in our community. So, I wrote an open letter in 2011 with my friends to highlight this wastefulness, and to encourage intelligent data archiving, and Nature News wrote a story about it which helped spread it around the palaeontology community. Since then, databases like https://morphobank.org/ have had a higher-rate of contribution but the general problem still remains: data is largely still a second-class citizen relative to written publications.
You were one of the first Panton Fellows. What does that mean, and how did this change your career?
Panton Fellowships are competitively awarded by the Open Knowledge Foundation to graduate and early career researchers, their goal is to empower the fellows promote open data in their research fields. Successful projects embrace the Panton Principles for open data in science, which in short recognise that:
Science is based on building on, reusing, and openly criticising the published body of scientific knowledge. For science to effectively function, and for society to reap the full benefits from scientific endeavours, it is crucial that science data be made open.
My fellowship gave me a strong sense of purpose to do something positive with my disillusionment of the way in which data was made available in my discipline. It was and still is, a real highlight of my CV. The recognition and financial and moral support of this award gave me the confidence to speak-up about open data related issues in science at many different conferences, bringing the issues to an audience of scientists that can be otherwise reluctant to listen to anything that isn't narrow, subject-related, academic research. The fellowship opened my eyes to the importance of policy-making & policy-influence, something commonly dismissed in traditional academia. Indeed, the most popular talk I have ever given was at a meeting of the European Commission (EC) 'Licenses for Europe' Text & Data Mining Working Group, in Brussels, providing evidence to the EC as to what challenges and difficulties European researchers face in this type of research. I would never have been at this meeting, or many like it, making a positive impact on research policy, if it wasn't for the Panton Fellowship.
The fellowship also changed the direction of my academic research. Together with one of the mentors of my fellowship award (Peter Murray-Rust) and my PhD supervisor (Matthew Wills), the three of us wrote a very open science-y grant proposal to liberate and make data buried in the literature re-usable again, which was successful and is what I'm working on now in my first postdoc, the PLUTo project.
What does open mean to you as a scientist, and how can other scientists be more open?
Take a formal definition of open, whether in the context of science or outside it, like the The Open Definition:
"A piece of data or content is open if anyone is free to use, reuse, and redistribute it—subject only, at most, to the requirement to attribute and/or share-alike."
In the context of science, this means that academic publications are only open access if they are licensed under OKD-compliant open licenses such as the Creative Commons Attribution Licence or the Creative Commons Zero Waiver. Likewise, data is only open data if it is explicitly licensed under an OKD-compliant licence, or otherwise clearly not subject to copyright.
It may seem tedious to be so precise about the definition of open but it really does matter. The figure images that I'm mining for the PLUTo project are clearly subject to copyright, even if they contain uncopyrightable data. I can re-post open-licensed figures of evolutionary trees on Flickr here which makes my research-process more accessible (less boring!) and searchable. I can get community-aided tagging of content and view metrics to demonstrate impact.
But research figures not published under open licenses don't/can't get this treatment, and I have a much larger collection of these currently languishing on my hard-drives. I'm simply not allowed to share them, even though the collection-as-a-whole if posted openly online would be far more useful to the community. Publisher-imposed restrictions mean I can only re-post perhaps 10% of the relevant figures I'm finding.
Scientists themselves have everything to gain from doing open scholarship, and there are some very simple steps that can be taken in that direction, namely: posting preprints and using your institutional or subject repository for all your research outputs (specifically including code and data, not just publications). Evidence shows there's a clear citation advantage for both open access publications and publications supplying open data, so it really is in the interest of the individual to do open scholarship.
Do you see scope for greater interaction with the open source community in the future?
Absolutely. Open source is clearly 'winning' now in my areas of science (ecology, palaeontology, phylogenetics). Open source software like R and programming languages like Python are extremely popular. Online platforms like GitHub are almost single-handedly transforming academic culture, getting many scientists to use proper distributed version-control systems for the first-time, through the use of git. I've even helped write an academic paper on GitHub! It's an extremely exciting time for open science and its intersection with the open source community.
View the complete collection of articles from Careers in Open Source Week.








Comments are closed.